
The emergence and spread of various multidrug-resistant bacterial strains have posed alarming challenges to public health and agriculture worldwide. Public demand for new antibiotics is enormous, yet drug development pipelines of the pharmaceutical industry started to run dry with limited targets available for inventing new bactericidal antibiotics. Our group has been working on molecular mechanism of virulence in model bacterial pathogens including Pseudomonas syringae, Pseudomonas aeruginosa, Bacillus cereus, Klebsiella pneumoniae. These pathogens rely on multiple system to invade their hosts, which is finely regulated by a group of transcription factors and signaling pathways. Research in our group has led to the identification of a variety of new virulence-associated two-component systems, transcription factors and their molecular mechanisms. Our lab has mapped the global transcription factor-based regulatory networks for these superbugs, and found a couple of potent lead compounds inhibiting them.
Pseudomonas syringae is a model organism for plant pathogenic bacteria, and is widely considered as the Top 1 plant bacterial pathogen, causing deadly diseases and huge economic lost in agriculture worldwide. During the past 14 years, we have screened a series of transposon-insertion libraries, and identified over 200 mutants displaying compromised expression of T3SS genes in P. syringae. Our previous studies demonstrate that a group of key regulators (such as AefR and RhpRS) and new regulatory mechanisms (such as Lon and HrpS) enable P. syringae to coordinate T3SS gene expression. Our long-term goal is to understand the missing signal(s) and overall signaling pathways that regulate T3SS, and leverage the outcomes of our studies to develop better therapies against P. syringae infection.
P. aeruginosa is one of the most commonly-isolated nosocomial pathogen, accounting for 10% of all hospital-acquired infections worldwide. In 2017, the World Health Organization (WHO) published its first ever list of antibiotic-resistant "priority pathogens" — 12 families of bacteria that pose the greatest threat to human health, and carbapenem-resistant P. aeruginosa was ranked as the Top 2 with critical priority. Recent research in my lab has led to the identification of a variety of new virulence factors of QS and their molecular mechanisms. Our long-term goals are directed at identifying new virulence factors and investigating the molecular pathogenesis in this bacterium. We plan to comprehensively understand it overall regulatory network, which will help to develop novel and effective approaches against P. aeruginosa infection.
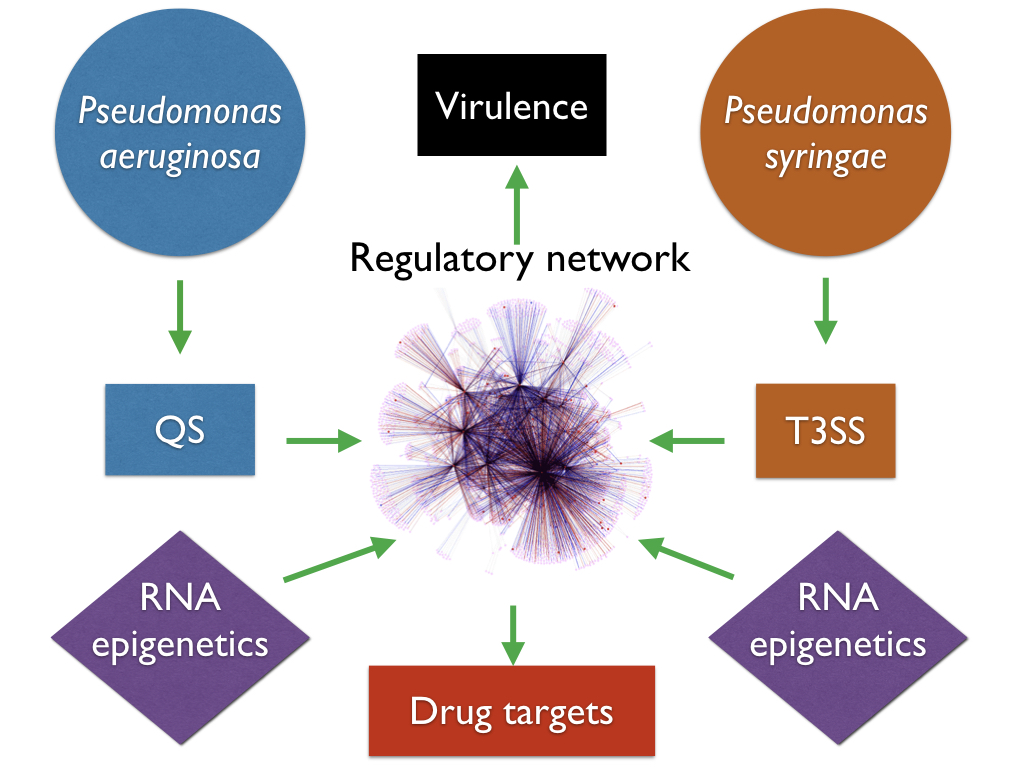
The other topic in my group is RNA epigenetics in bacteria. We have mapped the m6A methylome and rG4 structures in bacteria, and found unique patterns other than eukaryotes. Our work demonstrates that m6A and rG4 exist in a wide-range of bacterial species, which suggests their potential important roles in gene regulation.